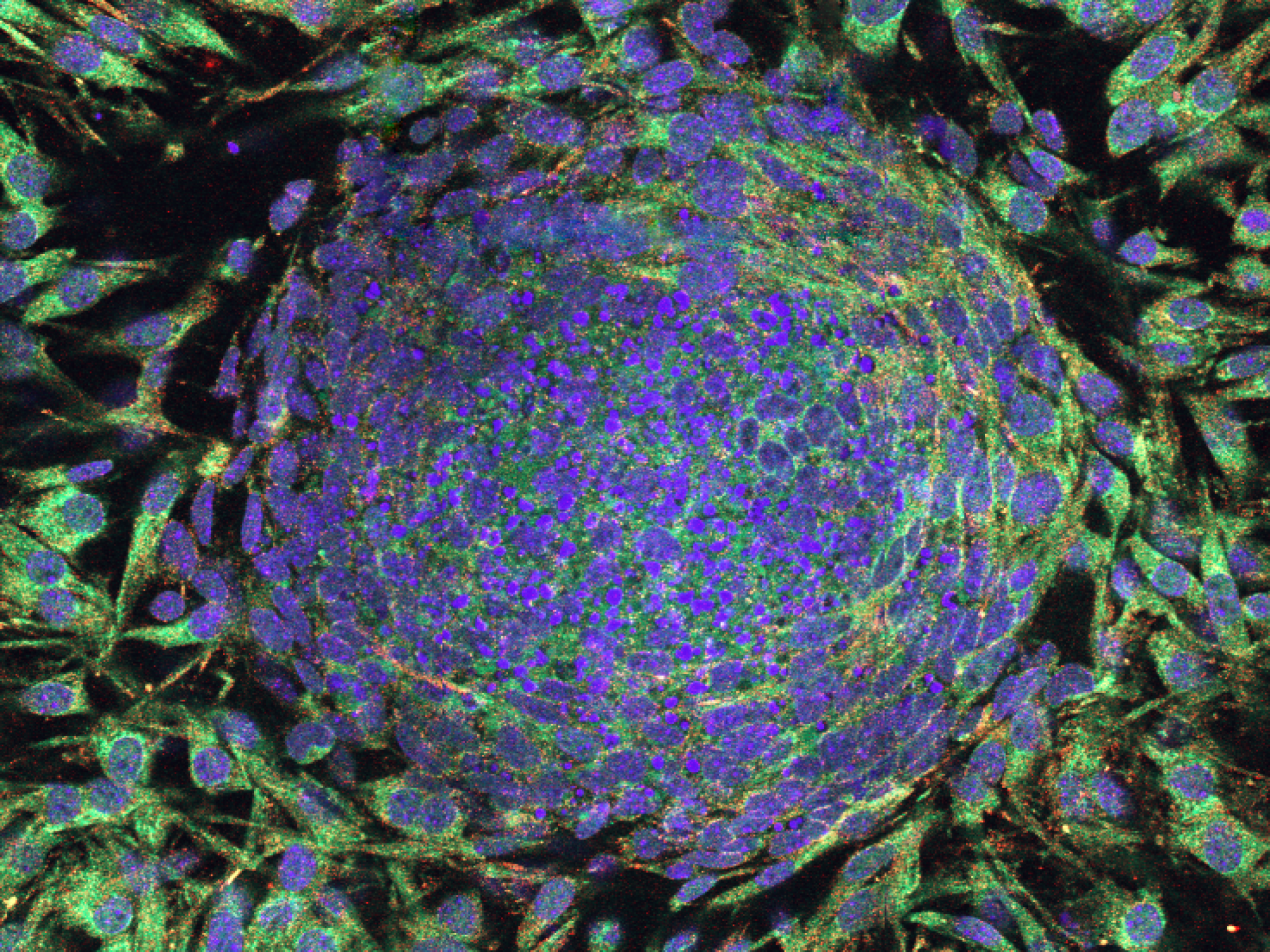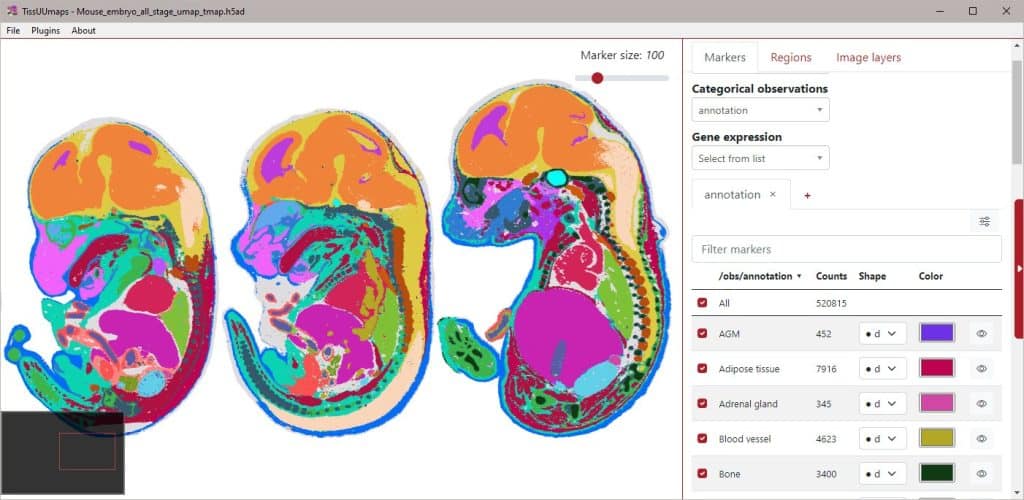
We present TissUUmaps, browser-based tool for GPU-accelerated visualization and interactive exploration of tens of millions of datapoints overlaying tissue samples. Users can visualize markers and regions, explore spatial statistics and quantitative analyses of tissue morphology, and assess the quality of decoding in situ transcriptomics data. TissUUmaps provides instant multi-resolution image viewing, can be customized, shared, and also integrated in Jupyter Notebooks. It is also possible to directly connect spatial markers with markers in feature space, such as UMAP plots, to interactively relate feature space with physical space. TissUUmaps was created in collaboration between BIIF and the Wählby lab. You can read more about it and test the software on its web page: https://tissuumaps.github.io/
During the seminar, we will specifically showcase new features of TissUUmaps 3.1, such as:
– HDF5 / AnnData files loading
– Network diagram visualization
– Multiple datasets displayed on a grid
– Plugin engine
The webinar will be given by Christophe Avenel and Fredrik Nysjö.
Organizer/NMI node: BIIF at Uppsala University
Location: Online
Time of event: 10:00-12:00, February 14th, 2023
Deadline for registration: February 14, 2023
Weblink: https://www.scilifelab.se/event/tissuumaps-3-interactive-visualization-and-quality-assessment-of-large-scale-spatial-omics-2/
Contact: christophe.avenel@it.uu.se